Laboratory Data
laboratory-data.RmdIntroduction
The article documents how to use the functions in
aeplots to analyse laboratory data in clinical trials.
Input data
2 treatment arms
To generate tables and plots to summarise laboratory data in clinical trials, the R package requires the input dataset in data frame format. A sample dataset with two treatment arms is shown below:
| id | arm | visit | lab_test | base | aval | lower | upper | region | strat | time |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Intervention | Week 0 | Lymphocytes (GI/L) | 2.57 | 2.57 | 0.85 | 4.1 | Other | < 1 year | 0 |
| 1 | Intervention | Week 4 | Lymphocytes (GI/L) | 2.57 | 1.87 | 0.85 | 4.1 | Other | < 1 year | 4 |
| 1 | Intervention | Week 8 | Lymphocytes (GI/L) | 2.57 | 3.14 | 0.85 | 4.1 | Other | < 1 year | 8 |
| 1 | Intervention | Week 12 | Lymphocytes (GI/L) | 2.57 | 1.72 | 0.85 | 4.1 | Other | < 1 year | 12 |
| 1 | Intervention | Week 16 | Lymphocytes (GI/L) | 2.57 | 2.09 | 0.85 | 4.1 | Other | < 1 year | 16 |
| 1 | Intervention | Week 20 | Lymphocytes (GI/L) | 2.57 | 2.22 | 0.85 | 4.1 | Other | < 1 year | 20 |
| 1 | Intervention | Week 24 | Lymphocytes (GI/L) | 2.57 | 2.00 | 0.85 | 4.1 | Other | < 1 year | 24 |
| 1 | Intervention | Week 0 | Monocytes (GI/L) | 0.49 | 0.49 | 0.20 | 1.1 | Other | < 1 year | 0 |
| 1 | Intervention | Week 4 | Monocytes (GI/L) | 0.49 | 0.46 | 0.20 | 1.1 | Other | < 1 year | 4 |
| 1 | Intervention | Week 8 | Monocytes (GI/L) | 0.49 | 0.42 | 0.20 | 1.1 | Other | < 1 year | 8 |
We would need to convert the visit column to ordered factor.
lab2$visit <- ordered(lab2$visit, c("Week 0", "Week 4", "Week 8", "Week 12", "Week 16", "Week 20", "Week 24"))Note that for all the functions below, you would need to specify the column names corresponding to each variable needed unless you rename the column names of your dataset to the default column names as specified in the sample dataset of the documentation. Do refer to the detailed documentation of each function by typing:
help(labtable)More than 2 treatment arms
The function labtable can take up to 4 treatment arms. A
sample dataset with 3 treatment arms:
| id | arm | visit | lab_test | base | aval | lower | upper | region | strat | time | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2570 | 135 | Placebo | Week 12 | Monocytes (GI/L) | 0.35 | 0.26 | 0.2 | 1.1 | Other | >= 1 year | 12 |
| 2571 | 135 | Placebo | Week 16 | Monocytes (GI/L) | 0.35 | 0.28 | 0.2 | 1.1 | Other | >= 1 year | 16 |
| 2572 | 135 | Placebo | Week 20 | Monocytes (GI/L) | 0.35 | 0.26 | 0.2 | 1.1 | Other | >= 1 year | 20 |
| 2573 | 135 | Placebo | Week 24 | Monocytes (GI/L) | 0.35 | 0.36 | 0.2 | 1.1 | Other | >= 1 year | 24 |
| 2574 | 135 | Placebo | Week 0 | Potassium (mmol/L) | 4.05 | 4.05 | 3.5 | 5.3 | Other | >= 1 year | 0 |
| 2575 | 135 | Placebo | Week 4 | Potassium (mmol/L) | 4.05 | 3.91 | 3.5 | 5.3 | Other | >= 1 year | 4 |
| 2576 | 135 | Placebo | Week 8 | Potassium (mmol/L) | 4.05 | 4.17 | 3.5 | 5.3 | Other | >= 1 year | 8 |
| 2577 | 135 | Placebo | Week 12 | Potassium (mmol/L) | 4.05 | 4.00 | 3.5 | 5.3 | Other | >= 1 year | 12 |
| 2578 | 135 | Placebo | Week 20 | Potassium (mmol/L) | 4.05 | 4.69 | 3.5 | 5.3 | Other | >= 1 year | 20 |
| 2579 | 135 | Placebo | Week 24 | Potassium (mmol/L) | 4.05 | 3.93 | 3.5 | 5.3 | Other | >= 1 year | 24 |
labtable function
2 treatment arms
labtable plots a table that summarises laboratory values
with continuous outcomes for baseline and each post-baseline timepoint
by treatment arms. It contains the mean & standard deviation (SD),
median & interquartile range (IQR), number of missing observations
and treatment effect estimate with its 95% confidence interval (CI).
Lab Measure |
Intervention (N₁=67) |
Placebo (N₂=68) |
Treatment effect estimate |
||
|---|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
2 (1.1) |
2 (0.9) |
0.0598 (-0.193, 0.313) |
Median (IQR) |
1.8 (1.1) |
1.9 (1.1) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
2 (0.9) |
1.9 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.8 (0.9) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
2 (1) |
1.9 (0.7) |
||
Median (IQR) |
1.9 (1.1) |
1.7 (0.8) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
2 (1) |
1.8 (0.7) |
||
Median (IQR) |
1.8 (0.8) |
1.8 (0.8) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
2 (0.9) |
2 (0.8) |
||
Median (IQR) |
1.7 (0.9) |
1.9 (1.1) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.9 (0.8) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
2 (0.8) |
1.9 (0.6) |
||
Median (IQR) |
1.8 (1.1) |
1.8 (1) |
|||
Number of missing |
3 |
5 |
|||
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.00342 (-0.0532, 0.0601) |
Median (IQR) |
0.4 (0.3) |
0.4 (0.2) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
0.5 (0.2) |
0.4 (0.2) |
||
Median (IQR) |
0.5 (0.2) |
0.4 (0.2) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.5 (0.2) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.3) |
0.4 (0.3) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.4 (0.3) |
|||
Number of missing |
3 |
5 |
|||
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
-0.031 (-0.119, 0.0572) |
Median (IQR) |
4.2 (0.4) |
4.2 (0.3) |
|||
Number of missing |
0 |
1 |
|||
Week 4 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.1 (0.5) |
4.2 (0.3) |
|||
Number of missing |
4 |
1 |
|||
Week 8 |
Mean (SD) |
4.2 (0.3) |
4.3 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.1 (0.4) |
|||
Number of missing |
1 |
3 |
|||
Week 12 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
3 |
5 |
|||
Week 16 |
Mean (SD) |
3.9 (NA) |
NA (NA) |
||
Median (IQR) |
3.9 (0) |
NA (NA) |
|||
Number of missing |
66 |
68 |
|||
Week 20 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.5) |
4.2 (0.4) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
2 |
7 |
We can specify the linear mixed model used to estimate the treatment
effect and 95% CI via the model_formula argument. The
default model is aval ~ arm + (1|id).
labtable(lab2, control="Placebo", intervention_levels=c("Intervention"), model_formula="aval ~ arm + base + strat + time + (1|id) + (1|region)")Lab Measure |
Intervention (N₁=67) |
Placebo (N₂=68) |
Treatment effect estimate |
||
|---|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
2 (1.1) |
2 (0.9) |
0.0646 (-0.0457, 0.175) |
Median (IQR) |
1.8 (1.1) |
1.9 (1.1) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
2 (0.9) |
1.9 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.8 (0.9) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
2 (1) |
1.9 (0.7) |
||
Median (IQR) |
1.9 (1.1) |
1.7 (0.8) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
2 (1) |
1.8 (0.7) |
||
Median (IQR) |
1.8 (0.8) |
1.8 (0.8) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
2 (0.9) |
2 (0.8) |
||
Median (IQR) |
1.7 (0.9) |
1.9 (1.1) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.9 (0.8) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
2 (0.8) |
1.9 (0.6) |
||
Median (IQR) |
1.8 (1.1) |
1.8 (1) |
|||
Number of missing |
3 |
5 |
|||
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
-0.00061 (-0.0302, 0.029) |
Median (IQR) |
0.4 (0.3) |
0.4 (0.2) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
0.5 (0.2) |
0.4 (0.2) |
||
Median (IQR) |
0.5 (0.2) |
0.4 (0.2) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.5 (0.2) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.3) |
0.4 (0.3) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.4 (0.3) |
|||
Number of missing |
3 |
5 |
|||
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
0.00101 (-0.0513, 0.0533) |
Median (IQR) |
4.2 (0.4) |
4.2 (0.3) |
|||
Number of missing |
0 |
1 |
|||
Week 4 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.1 (0.5) |
4.2 (0.3) |
|||
Number of missing |
4 |
1 |
|||
Week 8 |
Mean (SD) |
4.2 (0.3) |
4.3 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.1 (0.4) |
|||
Number of missing |
1 |
3 |
|||
Week 12 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
3 |
5 |
|||
Week 16 |
Mean (SD) |
3.9 (NA) |
NA (NA) |
||
Median (IQR) |
3.9 (0) |
NA (NA) |
|||
Number of missing |
66 |
68 |
|||
Week 20 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.5) |
4.2 (0.4) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
2 |
7 |
We can specify the number of decimal places for the mean, SD, median
and IQR columns as well as the number of significant figures for the
treatment effect estimate and 95% CI. We can also specify whether to
present IQR as a single “iqr” value or as 25th and 75th “percentile”
through the IQR_format argument.
labtable(lab2, control="Placebo", intervention_levels=c("Intervention"), IQR_format="percentile", mean_dp=2, SD_dp=2, median_dp=2,
IQR_dp=2, estimate_sf=4, CI_sf=4)Lab Measure |
Intervention (N₁=67) |
Placebo (N₂=68) |
Treatment effect estimate |
||
|---|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
2 (1.08) |
2.01 (0.85) |
0.05978 (-0.1933, 0.3129) |
Median (IQR) |
1.75 (1.33, 2.43) |
1.92 (1.32, 2.42) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
1.96 (0.94) |
1.9 (0.7) |
||
Median (IQR) |
1.74 (1.41, 2.5) |
1.77 (1.46, 2.3) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
2.04 (1.03) |
1.86 (0.75) |
||
Median (IQR) |
1.9 (1.38, 2.44) |
1.74 (1.37, 2.15) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
2.03 (1) |
1.84 (0.68) |
||
Median (IQR) |
1.79 (1.47, 2.3) |
1.79 (1.43, 2.2) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
1.95 (0.89) |
2 (0.82) |
||
Median (IQR) |
1.71 (1.43, 2.34) |
1.9 (1.38, 2.4) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
1.85 (0.84) |
1.95 (0.72) |
||
Median (IQR) |
1.75 (1.21, 2.27) |
1.89 (1.49, 2.29) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
1.95 (0.81) |
1.87 (0.65) |
||
Median (IQR) |
1.83 (1.32, 2.38) |
1.8 (1.42, 2.34) |
|||
Number of missing |
3 |
5 |
|||
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.48 (0.2) |
0.46 (0.19) |
0.003415 (-0.05324, 0.06011) |
Median (IQR) |
0.45 (0.34, 0.6) |
0.43 (0.35, 0.54) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
0.49 (0.18) |
0.44 (0.18) |
||
Median (IQR) |
0.48 (0.36, 0.57) |
0.42 (0.31, 0.52) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
0.48 (0.17) |
0.49 (0.23) |
||
Median (IQR) |
0.43 (0.36, 0.55) |
0.48 (0.35, 0.57) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
0.45 (0.18) |
0.47 (0.21) |
||
Median (IQR) |
0.45 (0.3, 0.56) |
0.44 (0.34, 0.6) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
0.49 (0.18) |
0.49 (0.22) |
||
Median (IQR) |
0.48 (0.34, 0.6) |
0.48 (0.33, 0.64) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
0.48 (0.19) |
0.47 (0.2) |
||
Median (IQR) |
0.46 (0.33, 0.61) |
0.46 (0.31, 0.61) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
0.47 (0.17) |
0.49 (0.23) |
||
Median (IQR) |
0.45 (0.36, 0.55) |
0.45 (0.3, 0.62) |
|||
Number of missing |
3 |
5 |
|||
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.17 (0.33) |
4.22 (0.29) |
-0.03097 (-0.1191, 0.05721) |
Median (IQR) |
4.16 (3.95, 4.36) |
4.15 (4.08, 4.39) |
|||
Number of missing |
0 |
1 |
|||
Week 4 |
Mean (SD) |
4.17 (0.35) |
4.22 (0.31) |
||
Median (IQR) |
4.12 (3.95, 4.37) |
4.21 (4.08, 4.38) |
|||
Number of missing |
4 |
1 |
|||
Week 8 |
Mean (SD) |
4.21 (0.31) |
4.26 (0.34) |
||
Median (IQR) |
4.2 (3.99, 4.42) |
4.14 (4.03, 4.46) |
|||
Number of missing |
1 |
3 |
|||
Week 12 |
Mean (SD) |
4.22 (0.31) |
4.23 (0.31) |
||
Median (IQR) |
4.2 (4.04, 4.4) |
4.24 (4.02, 4.4) |
|||
Number of missing |
3 |
5 |
|||
Week 16 |
Mean (SD) |
3.9 (NA) |
NA (NA) |
||
Median (IQR) |
3.9 (3.9, 3.9) |
NA (NA) |
|||
Number of missing |
66 |
68 |
|||
Week 20 |
Mean (SD) |
4.18 (0.33) |
4.22 (0.29) |
||
Median (IQR) |
4.19 (3.94, 4.42) |
4.2 (4.03, 4.44) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
4.2 (0.33) |
4.21 (0.31) |
||
Median (IQR) |
4.16 (3.98, 4.43) |
4.2 (3.98, 4.39) |
|||
Number of missing |
2 |
7 |
We can choose to drop either the Treatment effect estimate (95% CI),
Mean (SD), Median (IQR) or Number of missing n by specifying
treatment_effect_estimate=FALSE, mean=FALSE,
median=FALSE or n_missing=FALSE.
labtable(lab2, control="Placebo", intervention_levels=c("Intervention"), treatment_effect_estimate=FALSE,
median=FALSE, n_missing=FALSE)Lab Measure |
Intervention (N₁=67) |
Placebo (N₂=68) |
||
|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
2 (1.1) |
2 (0.9) |
Week 4 |
Mean (SD) |
2 (0.9) |
1.9 (0.7) |
|
Week 8 |
Mean (SD) |
2 (1) |
1.9 (0.7) |
|
Week 12 |
Mean (SD) |
2 (1) |
1.8 (0.7) |
|
Week 16 |
Mean (SD) |
2 (0.9) |
2 (0.8) |
|
Week 20 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
|
Week 24 |
Mean (SD) |
2 (0.8) |
1.9 (0.6) |
|
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
Week 4 |
Mean (SD) |
0.5 (0.2) |
0.4 (0.2) |
|
Week 8 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
|
Week 12 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
|
Week 16 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
|
Week 20 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
|
Week 24 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
|
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
Week 4 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
|
Week 8 |
Mean (SD) |
4.2 (0.3) |
4.3 (0.3) |
|
Week 12 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
|
Week 16 |
Mean (SD) |
3.9 (NA) |
NA (NA) |
|
Week 20 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
|
Week 24 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
More than 2 treatment arms
labtable does not include the treatment effect estimates
and their 95% CIs for datasets with more than 2 treatment arms. To
change the labels of control and interventions in the table, specify the
label for control in the argument control_name and specify
the intervention labels in the argument intervention_names.
Note that intervention_names should be specified using the same order as
the interventions specified in intervention_levels.
labtable(lab3, control="Placebo", control_name="No Drug",
intervention_levels=c("Intervention 1", "Intervention 2"), intervention_names=c("Drug A", "Drug B"))Lab Measure |
Drug A (N₁=44) |
Drug B (N₂=45) |
No Drug (N₃=46) |
||
|---|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
2 (0.8) |
Median (IQR) |
1.9 (0.9) |
1.8 (0.8) |
2 (0.8) |
||
Number of missing |
0 |
2 |
1 |
||
Week 4 |
Mean (SD) |
2 (1.1) |
1.9 (0.7) |
2 (0.9) |
|
Median (IQR) |
1.7 (0.9) |
1.7 (1) |
1.9 (1.1) |
||
Number of missing |
3 |
1 |
4 |
||
Week 8 |
Mean (SD) |
1.9 (0.9) |
2 (0.6) |
2.1 (1) |
|
Median (IQR) |
1.7 (1.1) |
1.8 (0.6) |
1.9 (0.8) |
||
Number of missing |
0 |
1 |
4 |
||
Week 12 |
Mean (SD) |
2 (0.9) |
1.9 (0.7) |
2 (0.7) |
|
Median (IQR) |
1.9 (1.3) |
1.9 (1.1) |
2 (0.7) |
||
Number of missing |
1 |
0 |
5 |
||
Week 16 |
Mean (SD) |
2 (0.8) |
1.9 (0.8) |
1.9 (0.8) |
|
Median (IQR) |
1.8 (1.3) |
1.8 (0.9) |
1.8 (0.6) |
||
Number of missing |
1 |
2 |
4 |
||
Week 20 |
Mean (SD) |
1.9 (0.8) |
2 (0.9) |
2 (0.8) |
|
Median (IQR) |
1.9 (1) |
1.9 (1.2) |
1.8 (1.3) |
||
Number of missing |
1 |
1 |
5 |
||
Week 24 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
1.9 (0.7) |
|
Median (IQR) |
1.8 (1.3) |
2 (0.9) |
1.9 (0.8) |
||
Number of missing |
3 |
0 |
5 |
||
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.5 (0.3) |
0.5 (0.2) |
0.4 (0.2) |
Median (IQR) |
0.4 (0.3) |
0.4 (0.2) |
0.4 (0.2) |
||
Number of missing |
0 |
2 |
1 |
||
Week 4 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.5 (0.2) |
|
Median (IQR) |
0.4 (0.3) |
0.5 (0.3) |
0.4 (0.2) |
||
Number of missing |
3 |
1 |
4 |
||
Week 8 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.5 (0.2) |
|
Median (IQR) |
0.5 (0.4) |
0.4 (0.2) |
0.5 (0.2) |
||
Number of missing |
0 |
1 |
4 |
||
Week 12 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.4 (0.2) |
|
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
0.4 (0.2) |
||
Number of missing |
1 |
0 |
5 |
||
Week 16 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.4 (0.1) |
|
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
0.4 (0.2) |
||
Number of missing |
1 |
2 |
4 |
||
Week 20 |
Mean (SD) |
0.5 (0.3) |
0.5 (0.2) |
0.5 (0.2) |
|
Median (IQR) |
0.5 (0.4) |
0.4 (0.2) |
0.5 (0.2) |
||
Number of missing |
1 |
1 |
5 |
||
Week 24 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.5 (0.2) |
|
Median (IQR) |
0.5 (0.3) |
0.5 (0.2) |
0.4 (0.2) |
||
Number of missing |
3 |
0 |
5 |
||
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.3 (0.3) |
4.2 (0.3) |
4.1 (0.3) |
Median (IQR) |
4.2 (0.4) |
4.2 (0.2) |
4.1 (0.4) |
||
Number of missing |
1 |
0 |
0 |
||
Week 4 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
4.1 (0.3) |
|
Median (IQR) |
4.2 (0.5) |
4.2 (0.4) |
4.2 (0.4) |
||
Number of missing |
2 |
1 |
2 |
||
Week 8 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
4.1 (0.3) |
|
Median (IQR) |
4.2 (0.4) |
4.2 (0.3) |
4.1 (0.3) |
||
Number of missing |
0 |
0 |
4 |
||
Week 12 |
Mean (SD) |
4.3 (0.3) |
4.3 (0.3) |
4.1 (0.3) |
|
Median (IQR) |
4.3 (0.3) |
4.2 (0.5) |
4.1 (0.3) |
||
Number of missing |
2 |
1 |
5 |
||
Week 16 |
Mean (SD) |
NA (NA) |
4.5 (NA) |
NA (NA) |
|
Median (IQR) |
NA (NA) |
4.5 (0) |
NA (NA) |
||
Number of missing |
44 |
44 |
46 |
||
Week 20 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
4.2 (0.3) |
|
Median (IQR) |
4.2 (0.4) |
4.3 (0.3) |
4.2 (0.4) |
||
Number of missing |
1 |
0 |
6 |
||
Week 24 |
Mean (SD) |
4.3 (0.3) |
4.2 (0.3) |
4.2 (0.3) |
|
Median (IQR) |
4.2 (0.3) |
4.2 (0.3) |
4.2 (0.4) |
||
Number of missing |
3 |
1 |
5 |
labscatter function
2 treatment arms
labscatter generates a scaterplot matrix to visualise
multiple continuous harm outcomes by treatment group. Specify the
laboratory tests of interest using the lab_test_list
argument. For each test, indicate whether the threshold represents an
upper or lower limit using the limit_list argument, and
provide the corresponding cutoff values for the dashed reference lines
using the cutoff_list argument.
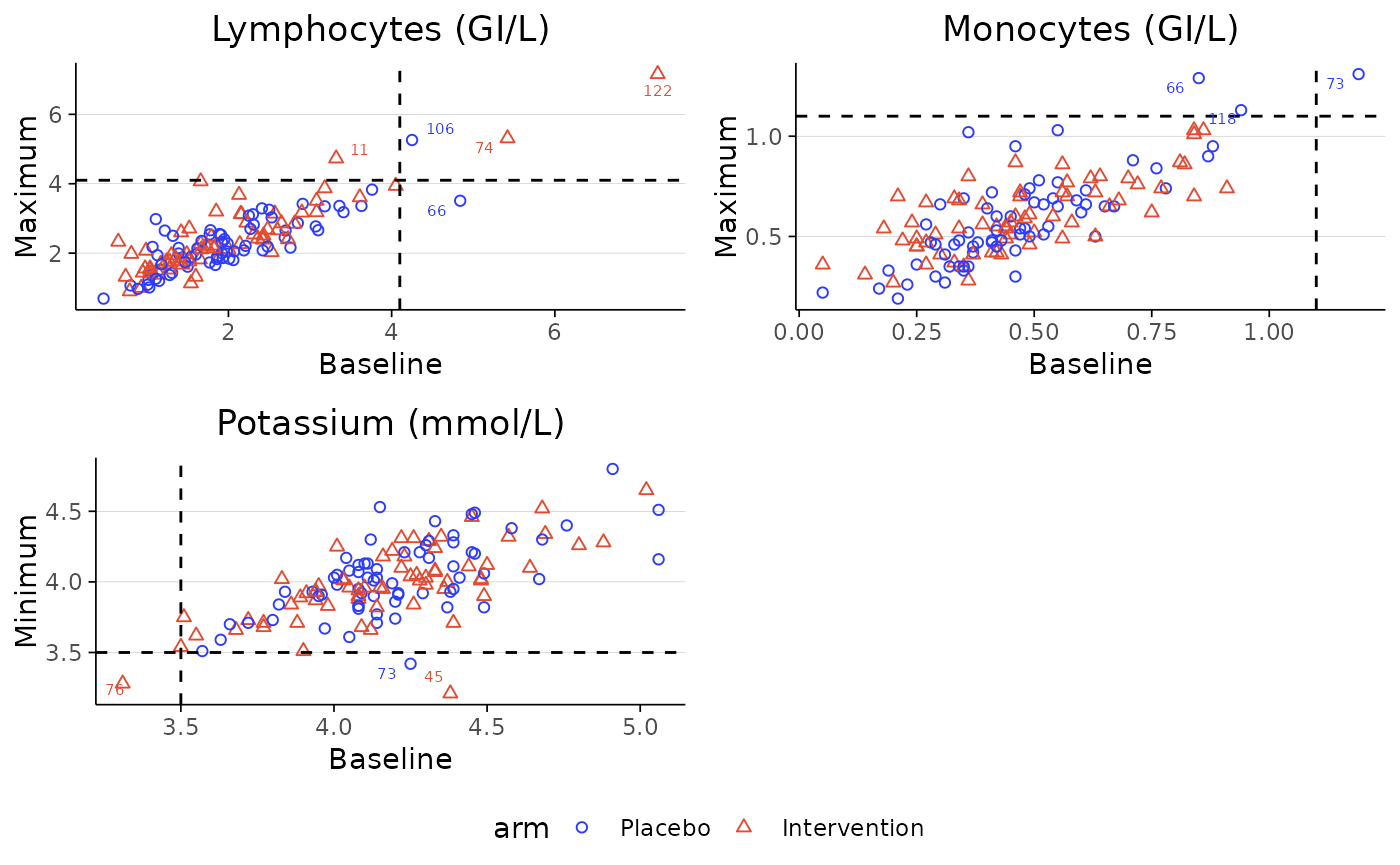
labscatter(lab2, arm_levels=c("Placebo", "Intervention"),
lab_test_list=c("Lymphocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)"), limit_list=c("upper", "upper", "lower"),
cutoff_list=c(4.1, 1.1, 3.5))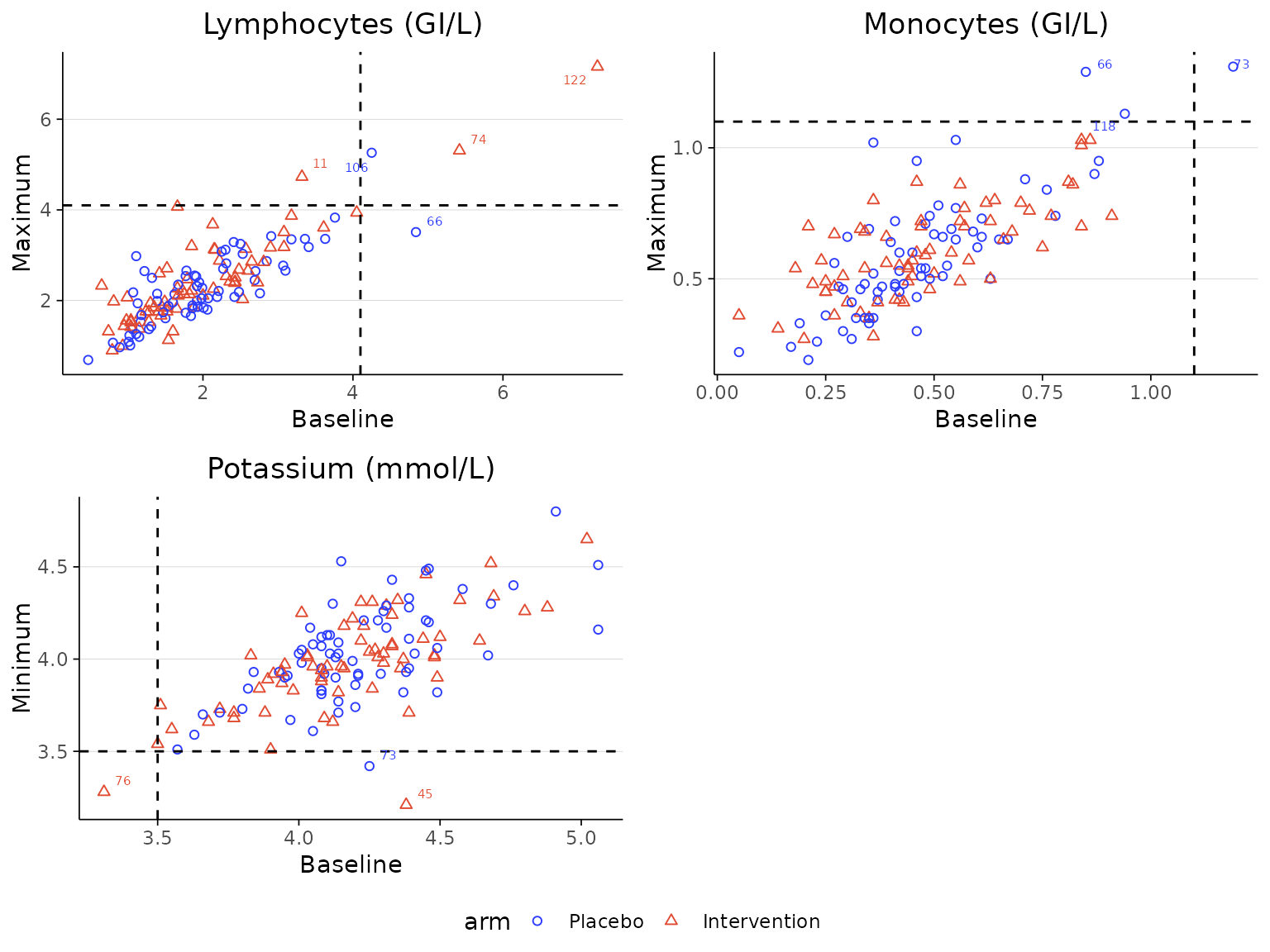
If the dataset contains lower and upper limits of the laboratory
measurements, the threshold lines can be drawn directly from the
lower and upper columns by specifying their
column names in the lower and upperarguments.
In this case, the cutoff_list argument does not need to be
specified.
labscatter(lab2, arm_levels=c("Placebo", "Intervention"),
lab_test_list=c("Monocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)", "Potassium (mmol/L)"),
limit_list=c("upper", "lower", "upper", "lower"),
lower="lower", upper="upper")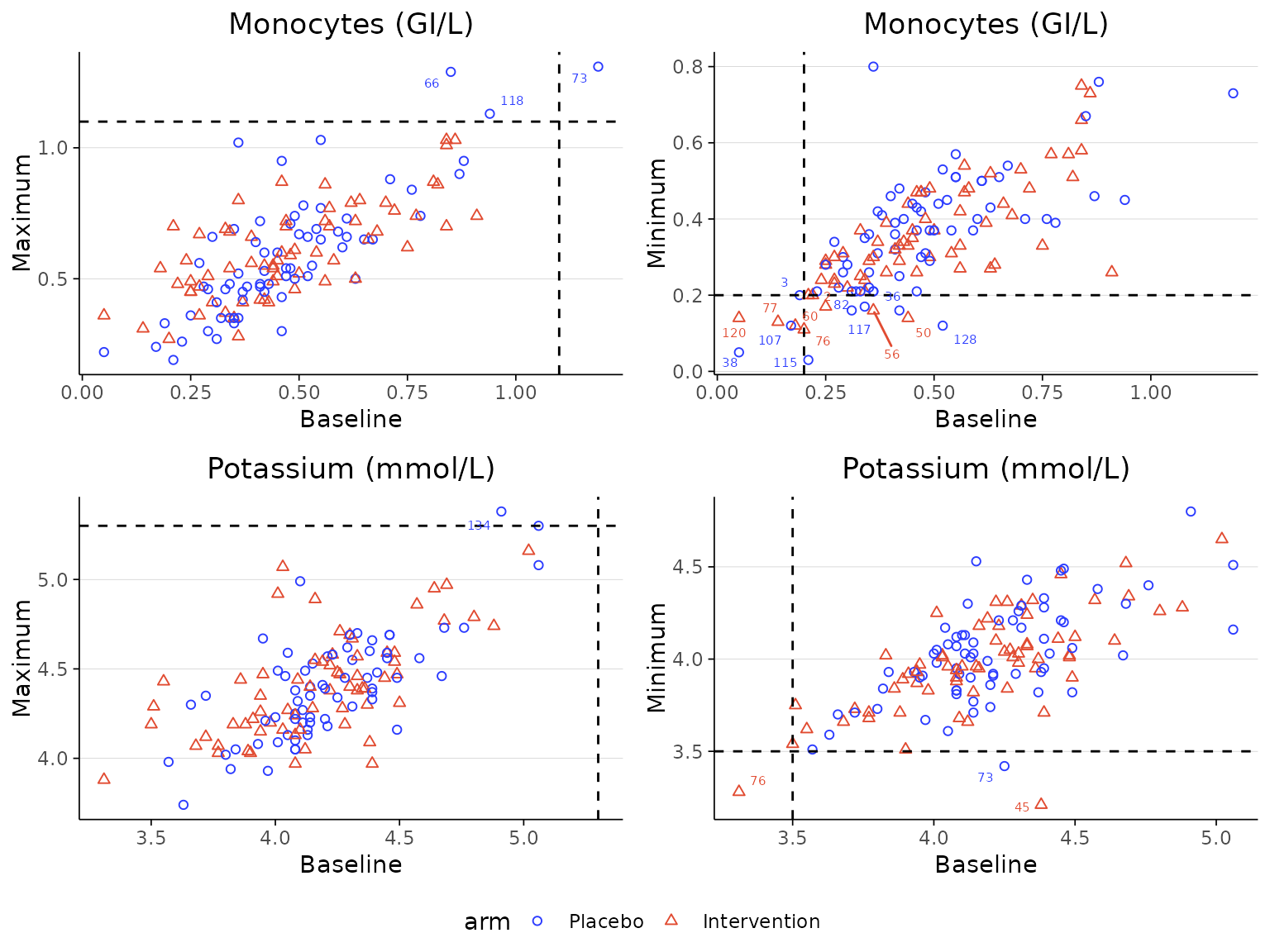
Instead of plotting the baseline value against the maximum/minimum
post-baseline laboratory measurement value, the plot can be configured
to use the final follow-up visit value by setting
last_visit=TRUE.
labscatter(lab2, arm_levels=c("Placebo", "Intervention"),
lab_test_list=c("Lymphocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)"), limit_list=c("upper", "upper", "lower"),
cutoff_list=c(4.1, 1.1, 3.5), last_visit=TRUE)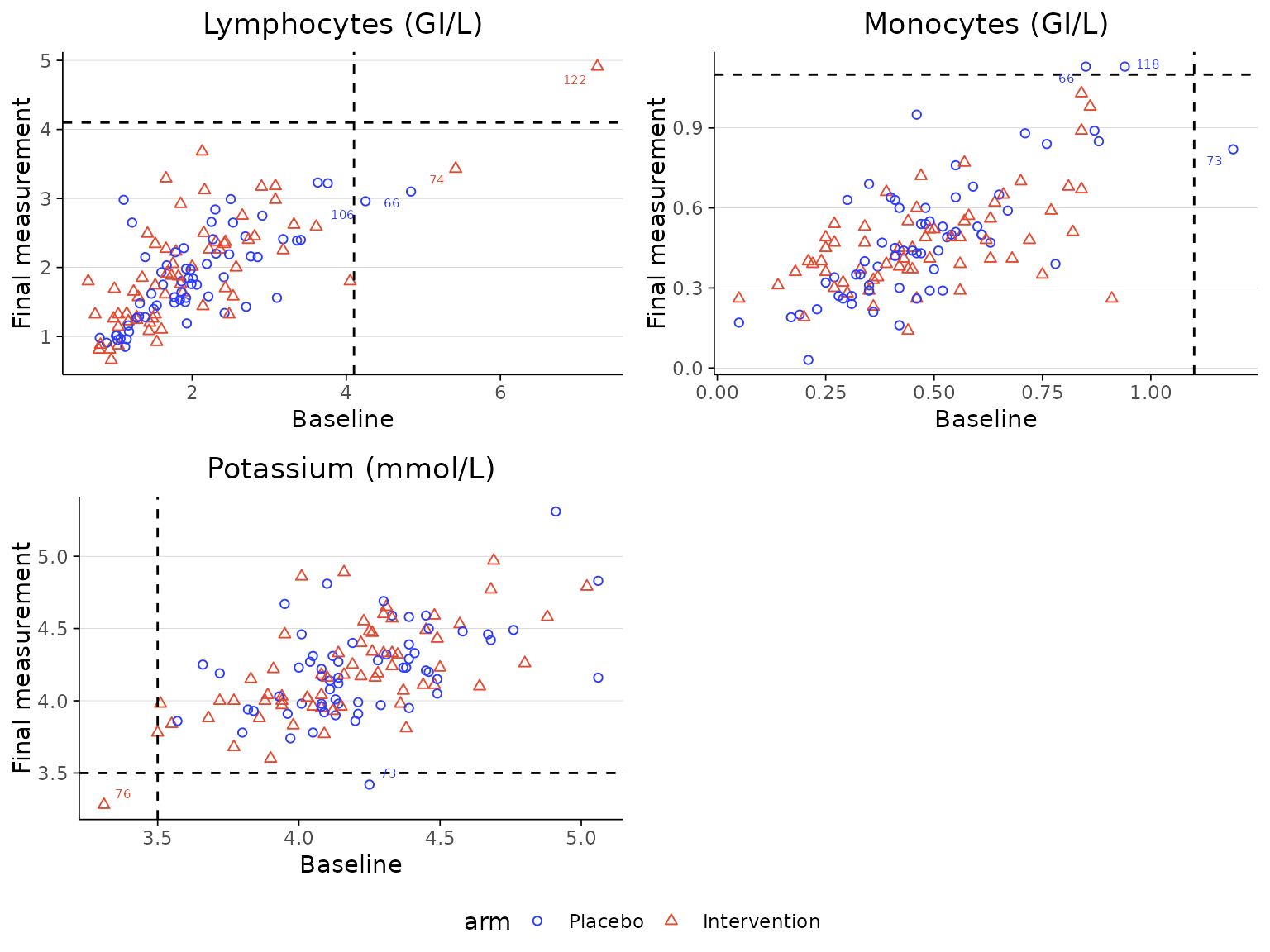
We can change the colour representing each treatment arm by
specifying a vector of colour codes in the arm_colours
argument according to the order of arm levels specified in the
arm_levels argument.
labscatter(lab2, arm_levels=c("Placebo", "Intervention"),
lab_test_list=c("Lymphocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)"), limit_list=c("upper", "upper", "lower"),
cutoff_list=c(4.1, 1.1, 3.5), arm_colours=c("#6AA84f", "#F1C232"))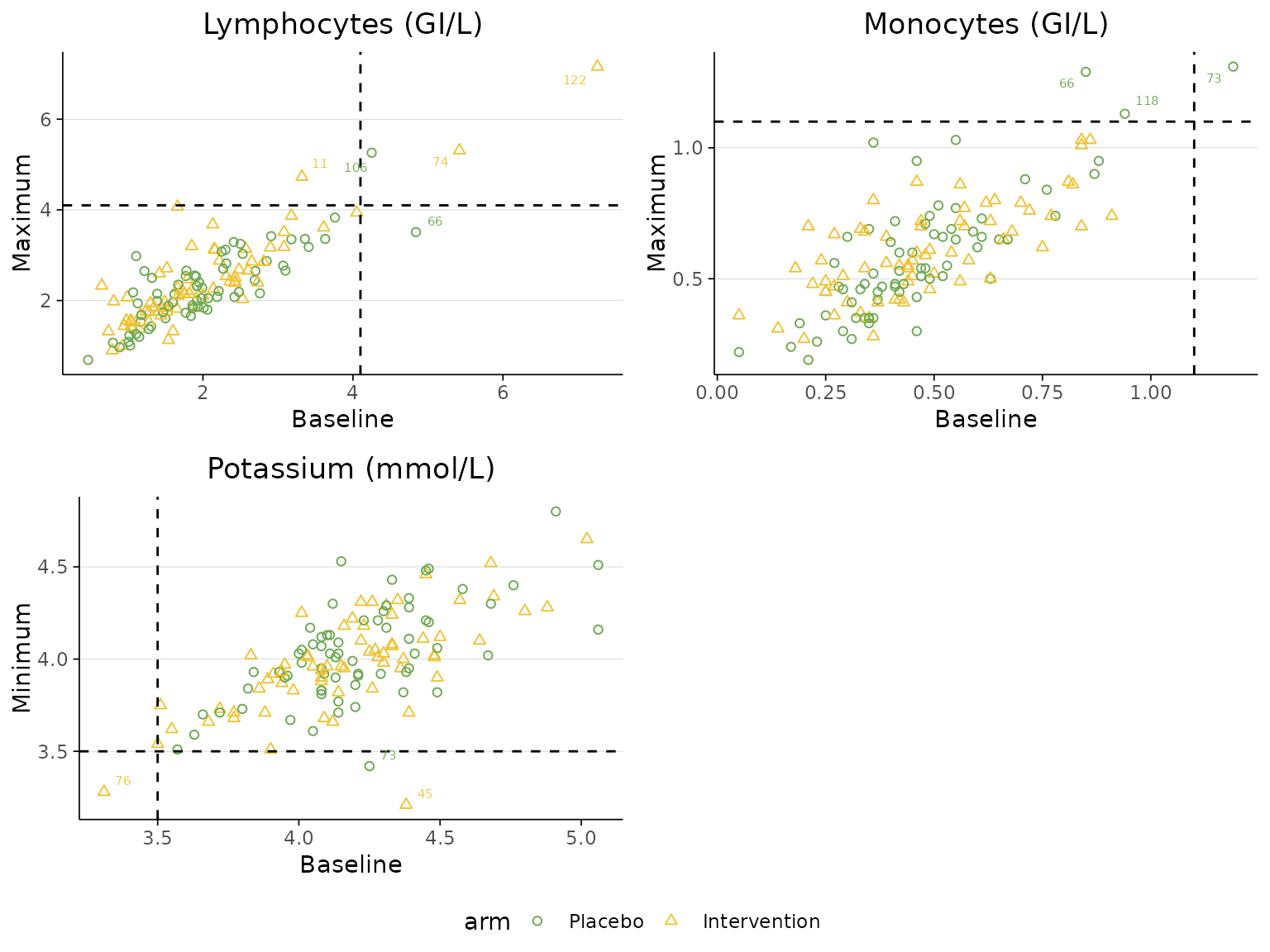
More than 2 treatment arms
labscatter(lab3, arm_levels=c("Placebo", "Intervention 1", "Intervention 2"),
lab_test_list=c("Lymphocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)"), limit_list=c("upper", "upper", "lower"),
cutoff_list=c(4.1, 1.1, 3.5))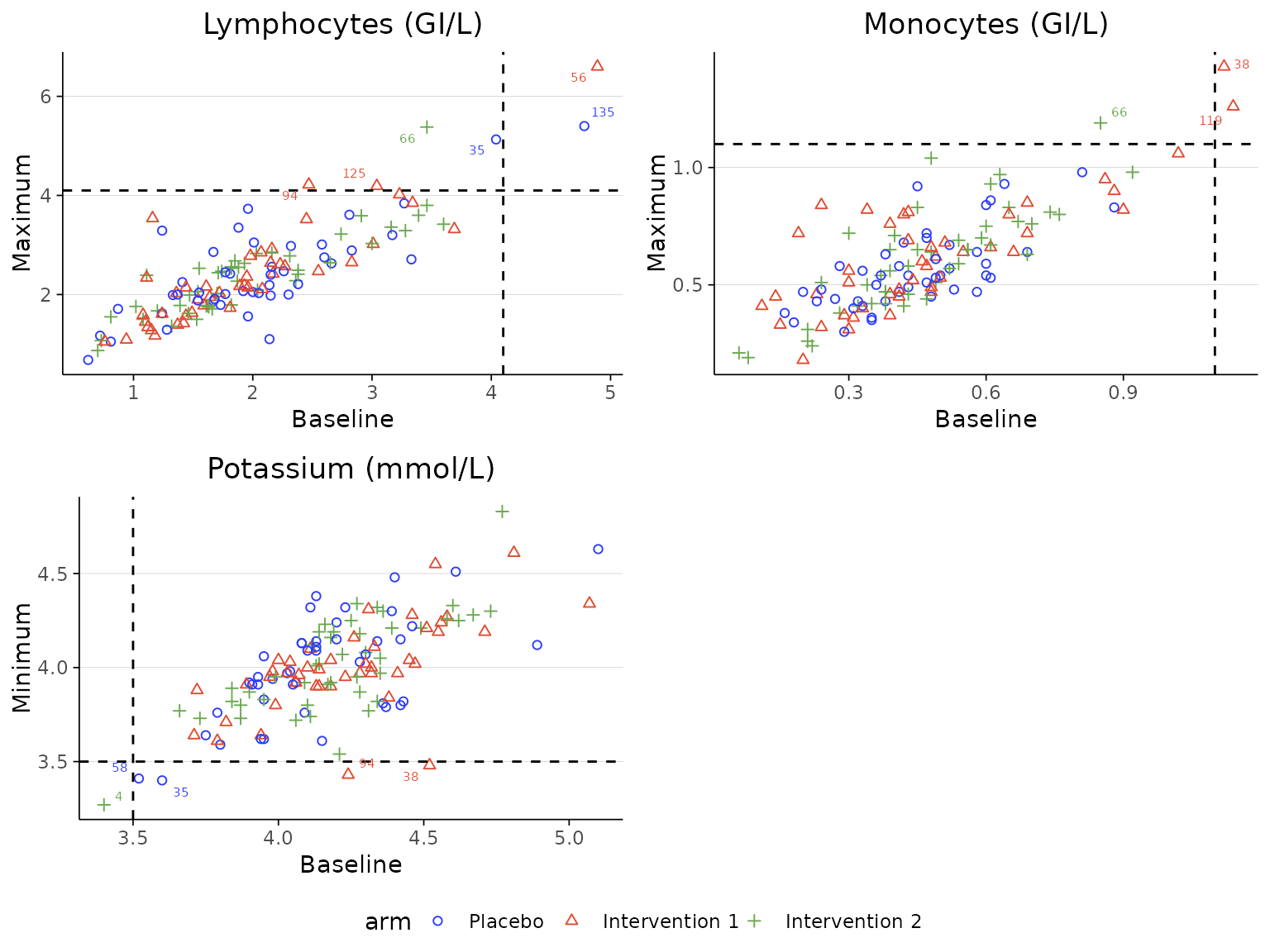
Saving tables and plots
Docx
We can save tables as docx by specifying the filepath in the
save_docx_path argument.
labtable(lab2, control="Placebo", intervention_levels=c("Intervention"), save_docx_path="labtable.docx")Lab Measure |
Intervention (N₁=67) |
Placebo (N₂=68) |
Treatment effect estimate |
||
|---|---|---|---|---|---|
Lymphocytes (GI/L) |
Week 0 |
Mean (SD) |
2 (1.1) |
2 (0.9) |
0.0598 (-0.193, 0.313) |
Median (IQR) |
1.8 (1.1) |
1.9 (1.1) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
2 (0.9) |
1.9 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.8 (0.9) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
2 (1) |
1.9 (0.7) |
||
Median (IQR) |
1.9 (1.1) |
1.7 (0.8) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
2 (1) |
1.8 (0.7) |
||
Median (IQR) |
1.8 (0.8) |
1.8 (0.8) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
2 (0.9) |
2 (0.8) |
||
Median (IQR) |
1.7 (0.9) |
1.9 (1.1) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
1.9 (0.8) |
2 (0.7) |
||
Median (IQR) |
1.7 (1.1) |
1.9 (0.8) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
2 (0.8) |
1.9 (0.6) |
||
Median (IQR) |
1.8 (1.1) |
1.8 (1) |
|||
Number of missing |
3 |
5 |
|||
Monocytes (GI/L) |
Week 0 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
0.00342 (-0.0532, 0.0601) |
Median (IQR) |
0.4 (0.3) |
0.4 (0.2) |
|||
Number of missing |
0 |
3 |
|||
Week 4 |
Mean (SD) |
0.5 (0.2) |
0.4 (0.2) |
||
Median (IQR) |
0.5 (0.2) |
0.4 (0.2) |
|||
Number of missing |
4 |
4 |
|||
Week 8 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.5 (0.2) |
|||
Number of missing |
2 |
3 |
|||
Week 12 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.3) |
0.4 (0.3) |
|||
Number of missing |
1 |
5 |
|||
Week 16 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
3 |
4 |
|||
Week 20 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.5 (0.3) |
0.5 (0.3) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
0.5 (0.2) |
0.5 (0.2) |
||
Median (IQR) |
0.4 (0.2) |
0.4 (0.3) |
|||
Number of missing |
3 |
5 |
|||
Potassium (mmol/L) |
Week 0 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
-0.031 (-0.119, 0.0572) |
Median (IQR) |
4.2 (0.4) |
4.2 (0.3) |
|||
Number of missing |
0 |
1 |
|||
Week 4 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.1 (0.5) |
4.2 (0.3) |
|||
Number of missing |
4 |
1 |
|||
Week 8 |
Mean (SD) |
4.2 (0.3) |
4.3 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.1 (0.4) |
|||
Number of missing |
1 |
3 |
|||
Week 12 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
3 |
5 |
|||
Week 16 |
Mean (SD) |
3.9 (NA) |
NA (NA) |
||
Median (IQR) |
3.9 (0) |
NA (NA) |
|||
Number of missing |
66 |
68 |
|||
Week 20 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.5) |
4.2 (0.4) |
|||
Number of missing |
1 |
6 |
|||
Week 24 |
Mean (SD) |
4.2 (0.3) |
4.2 (0.3) |
||
Median (IQR) |
4.2 (0.4) |
4.2 (0.4) |
|||
Number of missing |
2 |
7 |
Image
We can save tables and plots as images by specifying the filepath in
the save_image_path argument.
labscatter(lab2, arm_levels=c("Placebo", "Intervention"),
lab_test_list=c("Lymphocytes (GI/L)", "Monocytes (GI/L)", "Potassium (mmol/L)"), limit_list=c("upper", "upper", "lower"),
cutoff_list=c(4.1, 1.1, 3.5), save_image_path="labimage.png")